Alumni

Jessica Shi
Bio
Jessica Shi is a rising senior at Jamestown High School. She passionately pursues both the fields of science and the arts, taking special interest in environmental science, biology, and mathematics. She challenges herself in the classroom by taking a rigorous load of APs and college classes and participates in various academic clubs at her school including Model UN, Envirothon, and Mu Alpha Theta. She loves to travel all around the world and is fluent in Mandarin and Spanish. She spends much of her week volunteering in her community for Key Club, Mathcounts, and her school. In her free time, she enjoys playing viola for her local youth orchestra, painting, spending time with friends, and playing tennis on her school team.
Project: Implementing a Visualizer for DNA Sequence Alignments
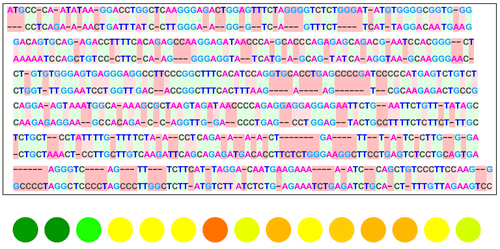
Goal
This project creates a visualization function that takes an input of a reference DNA sequence and multiple comparison sequences and outputs several different visual models that show the similarities and differences between them. It utilizes the functions SequenceAlignment, LongestCommonSubsequence, NeedlemanWunschSimilarity and SmithWatermanSimilarity, which are built-in functions of the Wolfram Language that use the classic algorithms for sequence alignment as created by the researchers Needleman, Wunsch, Smith and Waterman. The result will yield figures and graphs to show the alignment . This tool can be used to study evolutionary origins of species, detect mutations and analyze other biological relationships.
Summary of Results
These tools can be used to quickly analyze many sequences in reference to one sequence. Organisms that share similar bases and have similar sequence lengths and high alignment scores are more likely to have similar structure and function for that particular gene. It provides a large variety of visuals including charts and figures to compare the sequences for different purposes. It takes into account short sequence/long sequence differentiation and other syntactical differences. Possible applications include education or biological analysis.
Future Work
In the future, I hope to implement a DNA to mRNA codon to protein translator. This could be useful for learning about DNA transcription and translation. I could also modify the algorithm so that the sequences are compared among each other instead to a single reference strand. This would use a multiple sequence alignment algorithm instead of a pairwise sequence alignment algorithm. Another possible application would be using the results of DNA analysis to automatically generate phylogenetic trees among organisms.